Personal Science Week 230727 COVID
Reviewing my latest Serimmune results and how to think about COVID
Be curious, but be skeptical. When you see an impressive new scientific claim, try to see how it applies to you. Those are some of the principles of Personal Science.
This week I report on updated results from my Serimmune antibody test and speculate about what we know about COVID-19 infections.
More than three years have passed since COVID-19 became a thing. Meanwhile just about everyone you know has been infected. That includes everyone in my family except, so far, me. What’s different about me? Is it just a matter of time?
What about Serimmune
As long-time Personal Science Week readers know, I’ve been participating in a five-year clinical study from Serimmune, a California-based startup developing technologies for “immune intelligence”, a simple at-home blood test intended to uncover a person’s antibody levels for dozens of diseases, including tick-borne diseases like Lyme as well as COVID-19. (PROTO.LIFE published my in-depth article about my experience, and you can see previous PS Week reports from January 2023 and October 2022).
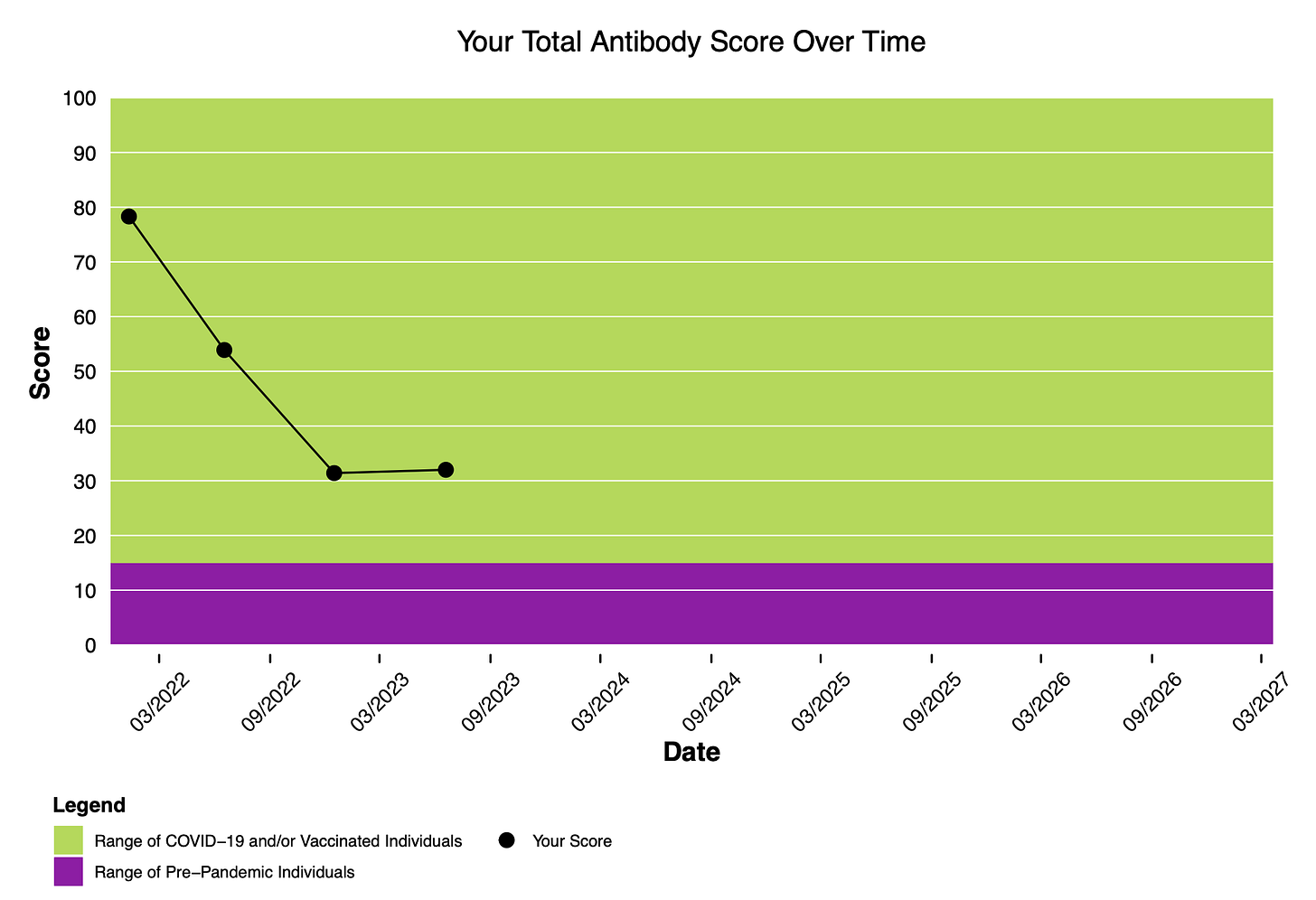
Study participants receive a sampling kit every six months, and my latest was in June 2023. As you can see, whatever COVID antibodies that Serimmune finds in me, the levels haven’t changed much in the past six months..
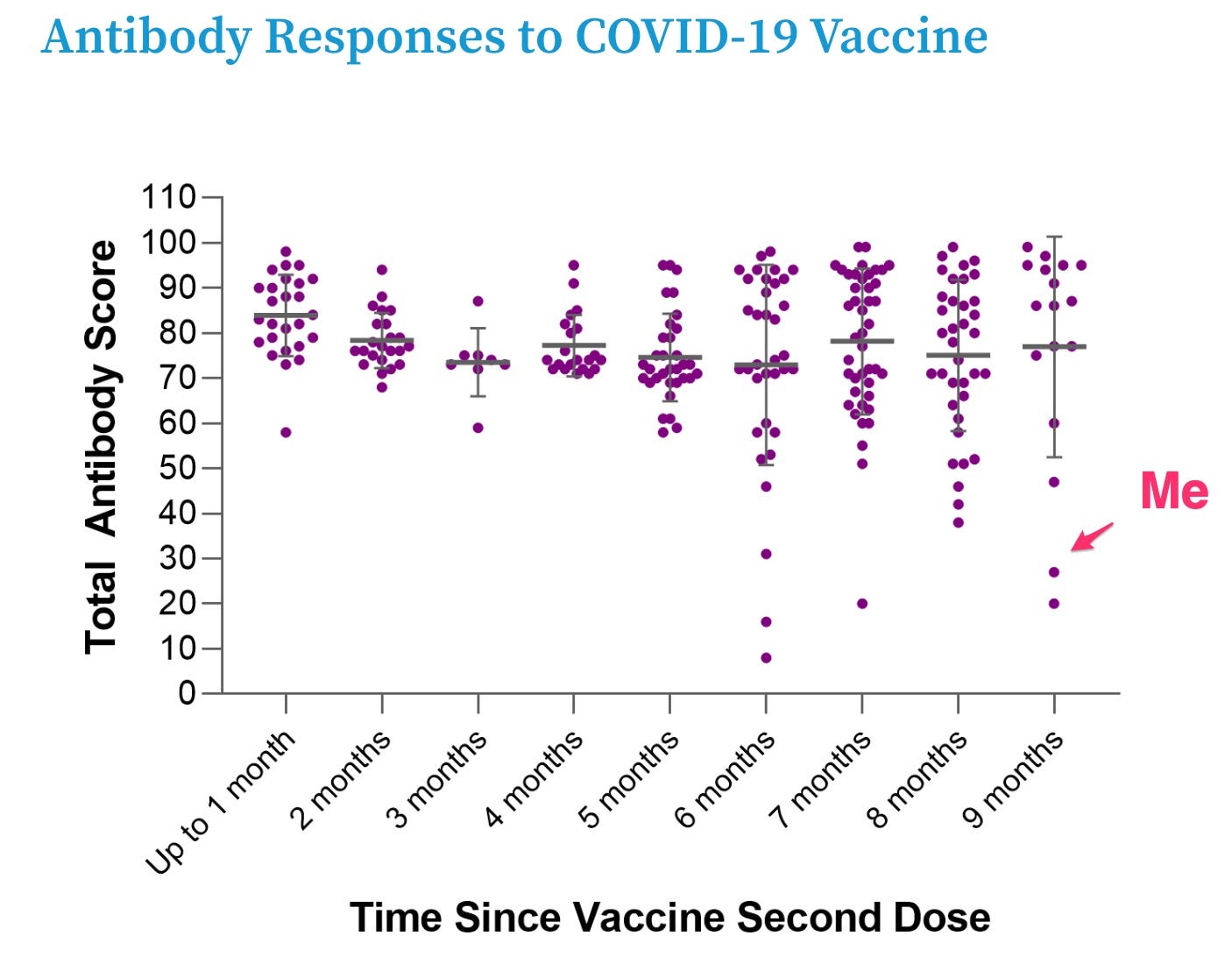
The company helpfully provides a summary of their study results so far that lets me compare my results to the other thousands of people in their study
Bottom line: my COVID antibodies are way lower than the other participants Serimmune has been studying.
Will I get COVID?
Here’s the thing: I’ve done nothing special to prevent an infection. For the past three years I’ve continued my day-to-day life as normal, never avoiding travel or meetings, and certainly not taking the draconian precautions of many of my more paranoid friends. My wife tested positive in May, after feeling a scratchy throat after a business trip in May. I was with her the whole time, no quarantining at all.
What’s especially odd to me is that it’s not just COVID. I haven’t had so much as a sniffle, not a cold, not a flu — nothing — since the pandemic began. This is probably the longest I’ve ever gone in my life without a cold.
How does COVID spread?
The surprising answer: nobody knows for sure. It probably has something to do with respiration, but that’s clearly not the whole story1. Look up the reports from the sailing ship Echizen Maru, where 57 crew members tested positive for covid-19 after more than a month of isolation on the high seas. Similarly, new cases popped up at the Belgian research station Princesse Elizabeth despite lengthy quarantines, multiple negative covid tests, and vaccination. Long before COVID, there was a documented case of Antarctic scientists catching cold after seventeen weeks of complete isolation. (see this Twitter thread for more). You can’t explain away these cases through a theory of person-to-person transmission.
The best way to confirm the mode of transmission would be through challenge trials: deliberately infect volunteers under carefully-controlled conditions. In fact, UK researchers recently tried this, giving SARS-CoV-2 virus laced nose drops to 36 healthy volunteers. Hard to imagine more of a direct way to infect somebody; volunteers were even isolated in special air pressure-controlled rooms. While about half of them became sick, half didn’t.
Something similar was tried during the Spanish Flu, when two challenge studies were attempted on 62 volunteers who were deliberately swabbed with active samples from sick patients. (Unfortunately?) the study was never published because nobody got sick.
And this conforms to your everyday experience as a Personal Scientist too. You personally know many cases of people who got it despite apparently doing everything “right”, while others in the same circumstances went unscathed. Professionals know less than you think about how viruses spread.
My One Preventative Strategy
Wait, it’s not quite true that I’ve done nothing to avoid COVID. I’ve made one behavior change since the beginning of the pandemic: I always, always, always wash my hands: before eating at a restaurant, throughout the day at work, as soon as I come home, and every other chance when it’s convenient.
No, I don’t use hand sanitizer, which I’m convinced is useless against viruses and can cause more harm than good by neutralizing, instead of killing germs2.

What about face masks? Because feelings are so strong one way or another, we’ll devote a future issue to this. But one conclusion is undeniable: if masks help, they don’t help very much. We all know people who wore (and still wear!) N95 face masks regularly and still became infected, often multiple times. They’ll always have some reasonable explanation (“I was very careful except that one time”).
I think when dealing with an unknown threat, you should try absolutely everything. But as a Personal Scientist, always be as self-critical as possible and ask yourself if it really works, or if you just think it does.
Genetics of COVID and Long COVID
Back in PS Week Oct 2022 we discussed Mentzer et al., 2022, a paper by Oxford scientists that looked at the genetics of a thousand people enrolled in a COVID vaccination trial. According to their preliminary data, people with HLA-DQB1*06 alleles were less likely to experience PCR-confirmed breakthrough infection. 23andme reports HLA-DQB1 status here. For me, it’s all Ts and Gs (no As). If I’m reading the paper correctly, that means I’m less likely to get a breakthrough infection. UPDATE 7/27 thanks to Julie Claire Green ND: I am indeed reading the paper incorrectly. Although the difference in infection rates is statistically significant, don’t get too excited: the overall difference is small, meaning you’ll likely be infected either way.
A new paper summarized in The Conversation (and Eric Topol summarizes the technicalities) describes a common variant of an HLA gene called HLA-B15:01* that is associated with asymptomatic infection. 3This variant is present in about 10% of the population with European ancestry.
Our findings suggest that preexposure to seasonal cold viruses allowed people with HLA-B15:01* to develop a very effective immune memory that helped them to quickly kill the virus before they developed symptoms.
Unfortunately, I know of no consumer test that can tell whether I have the alleles or not. The paper gets its data from blood samples taken from bone marrow transplant donors, and I’m told the HLA immunity-related genes involved are notoriously difficult to pin down in an over-the-counter genetic test.
Similarly, the COVID-19 host genetics initiative points to rs9367106 as a SNP associated with Long COVID, but unfortunately none of the other alleles mentioned are reported by 23andme (see my tweet).
About Personal Science
Nullius in verba — take no one’s word for it. We call it Personal Science, and this newsletter tries to be a brief, weekly summary of the consequences of that approach, hopefully in a way that will be interesting to others who are just as curious as we are. Paid subscribers have access to our series on Unpopular Science, ideas that are too controversial for today’s censorship-prone society.
As always, please let us know if you have other topics you’d like discussed
Links and References
The pseudonymous Substack writer eugyppius writes his theory of The Mysterious Disappearance of Influenza, speculating an understudied phenomenon of “viral interference” may explain some of the oddities of the pandemic.
Retired biochemist Stephen Andrews New hypothesis on the mechanism of SarsCov2 virus transmission is a detailed summary of the case for non-respiratory transmission.
Follow the excellent Dr. Annie’s detailed experiments to understand when and how hand sanitizers work. It’s part of her wonderful resource on how to stop the stomach flu.
That big study on alleles associated with immunity comes from the Covid Citizen Science Study (CCS), which you can join too.

I'm assuming "time since vaccine second dose" in their chart should read "time since last vaccine dose"?
It's going to be interesting to see if/how genetics can predict who is susceptible to (symptomatic) COVID. Might depend on the variant, too: I know someone whose spouse got COVID twice, but they only got infected after a lunch meeting with a friend... For those who are susceptible (i.e. most of us), the question isn't so much "will doing x ensure that I never get COVID", but whether doing x will a. delay getting infected (this was more important early on when health outcomes were worse), b. reduce the severity of an infection (outcomes appeared to be worse for people who were exposed to larger quantities of virus), and c. reduce the frequency of getting infected.
I would like to respectfully point out an error you made in the interpretation of this paper.
You mention "According to their preliminary data, people with HLA-DQB1*06 alleles were less likely to experience PCR-confirmed breakthrough infection. 23andme reports HLA-DQB1 status here. For me, it’s all Ts and Gs (no As). “
In the paper they do say “This AA variant” but that is not alleles AA, but amino acid.
HLA alleles were imputed from genotype data and the two alleles best tagging HLADQ B*6 were rs9271374 Glu71Arg (position 32661752, Chr 6, build 38) and rs1130456 DQB1-125A/S (Position 32659602, chr 6, Build 38). Neither of these SNPs appear to be in my raw data file of 23andme, version 3. Maybe they are in other versions. In essence you need to look at your specific results for those 2 SNPs (which can be triallelic…either A, C or T). HLA-DQB1*06 has a glycine at position 125, whereas other alleles common in the genotyped population possess either alanine (HLA-DQB1*02 and *04 alleles) or serine (HLA-DQB1*05). Thus, this amino acid variant (rs1130456 with Gly) is synonymous with the presence of HLA-DQB1*06 in our dataset.
These two rsid#s are not present (searching by position or rsid#). Sorry, once I found out these not in my raw data, I did not take the time to sleuth out what allele corresponds to Gly, Ala or Ser. Long story short: you cannot determine your HLA DQB1 status from 23andme, v3.
From the paper
These are SNPs located within 10 kilobases (kb) of HLA-DQ genes (Fig. 2) and in linkage disequilibrium within our multi-ancestry cohort (r 2 = 0.65). The distribution of P values (Extended Data Fig. 3a) and beta coefficients (Extended Data Fig. 3b) for all genotyped and imputed variants across this locus show a clear correlation in genetic architecture between these two antibody responses (Spearman’s rho coefficient 0.90 and 0.93 for P values and beta coefficients, respectively) correlated through linkage disequilibrium (measured through r 2 ).
HLA imputation was performed using the Multi-Ethnic HLA reference panel (version 1.0 2021) available on the Michigan Imputation Server46 using recommended settings. Phasing of multi-allelic HLA alleles was undertaken using PHASE (version 2.1.1)47. HLA typing was also performed using PCR sequence-specific primers (SSPs) at the Histogenetic laboratory, Oxford University NHS Foundation Trust.
This amino acid AA variant (DQB1-125A/S) denotes the presence of either an alanine or a serine at position 125 of the HLA-DQB1 protein according to international ImMunoGeneTics (IMGT) project coordinates. The index-associated variant from the primary analysis (rs1130456) was equally associated with the anti-RBD titers in this analysis (beta = 0.27 and s.e. = 0.04). Other variants imputed using the specific HLA imputation algorithm were identified as being marginally more significantly associated than rs1130456, with the new lead being rs9273817 (P = 2.4 × 10−9, beta = 0.27 and s.e. = 0.04).