How to think about microbiome diversity
Comparing multiple tests shows diversity is complicated
Most microbiome discussions begin with the assumption that diversity is good. After all, if your body harbors a wide variety of microbes, you’ll have a deeper catalog of useful ones that can be applied to new situations. The world around us is constantly changing, and you never know what new threats or opportunities you may encounter. You can respond better if you have an abundant variety of organisms that can meet any challenge.
In practice, diversity is difficult to pin down quantitatively. We know what we mean in principle — a rich variety of different microbes ought to be good — but clearly there are limits. You wouldn’t want “variety” to include serious pathogens, for example. We know intuitively that a deciduous forest at sea level, with dozens of different tree species, is more diverse than one at a high altitude tree line. But is the one at low altitudes “better”? It depends on whether you’re a squirrel or a mountain goat!
All diversity metrics take into account two aspects of a community: the number of different organisms in a sample, and the range of abundances for each one. To understand how this works, think of two forests, each with an equal number of trees.
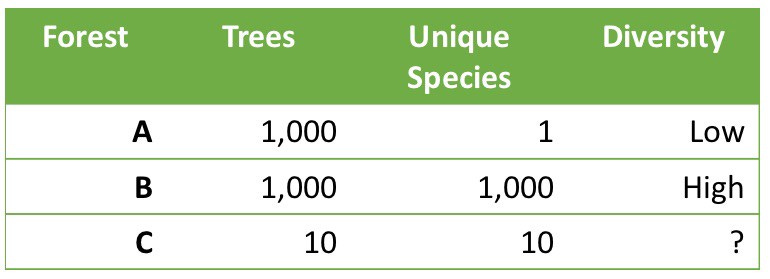
Which forest is better?
Clearly, Forest B with its abundance of species and trees is the most diverse. But what about Forest A compared to Forest C?
On the one hand, Forest C seems to have a greater variety of trees: 10 times more than Forest A. But it also has many fewer of them. In other words, there are two aspects of diversity that matter: the absolute number of organisms in an ecosystems, and the variety or richness of those that are there.
Whether A is “better” or “worse” than C depends on subjective, non-quantifiable factors that are not included in any diversity metric. A managed forest, such as one on a Christmas tree farm, might be perfectly healthy for one purpose (growing Christmas trees for sale), while an adjacent clear-cut forest with ten lonely and scraggly trees could be far less healthy, even if it has more of a variety of trees.
In this example, we use the term richness to refer to the situations in Forests B or C, with their greater variety of species. The broader term diversity tries to be a measure of both richness and abundance.
The microbiome testing company Viome (which I reviewed recently for NEO.LIFE) uses these terms to show how my microbiome compares to other people:
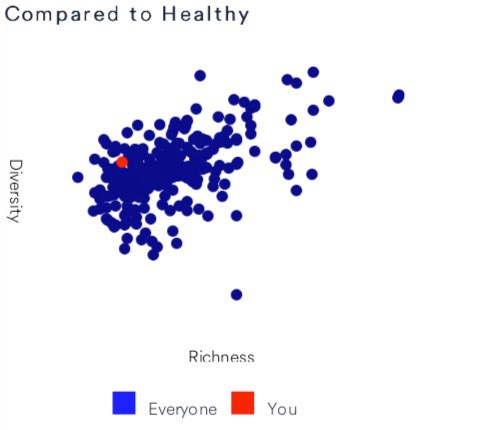
Here’s how the microbiome testing company Viome rates my diversity. This was taken on the same date, from the same sample as the one shown below for uBiome.
Viome, like other microbiome testing labs, generates this chart from the taxonomy tables they sequence, using the same principle described with the forest example above. A microbiome sample with 100 unique taxa (i.e. organisms at a particular taxonomic rank) is more diverse than one with only 10 unique taxa.
Quantifying diversity
But if we just use raw, absolute numbers, it can be hard to compare across different microbiome tests. For example, what if I have two samples, each with 100 unique taxa, but in one sample there are tiny amounts of all but one of the taxa, while the other sample has equal amounts of everything? Which is more diverse?
One way to quantify this is with a metric borrowed from probability theory. What if, instead of looking at all the taxa and their respective amounts, we simply take at random any two taxa from the sample: what is the probability that the two chosen will be the same?

Diversity measurements can be random. [Image: SalFalko]
If I have a sample with all unique taxa, each of identical abundance, then the odds are pretty low that I would select at random two of the same kind; conversely, if a majority of the sample consists of the same taxa, with many other taxa of smaller abundance, then the odds are pretty good that the two I select would be the same.
In fact this is generally the case in healthy western guts, which are usually composed of only two large phyla: Firmicutes and Bacteroidetes. In my case, these two phyla make up over 90% of everything I see in my samples, so the odds that you would randomly pick these two is pretty high. That’s the intuition behind the Inverse Simpson metric, developed in 1949 by the British scientist E.H. Simpson.
Another microbiome testing company, uBiome, tries to normalize the Inverse Simpson number on a scale from 1 to 10, which they then compare to all the other samples in their database. When I sent them a gut sample taken on the same day as the one above, uBiome said that my microbiome was more diverse than 89% of the others in their 250K+ sample database. Although Viome doesn’t explain how they calculated the chart above, they seem to peg me at about the middle of the scale compared to their healthy users.

Using the same sample as the one submitted to Viome, here’s how uBiome ranks the diversity of my gut microbiome, on a scale from one to ten.
The taxonomy of microbes matters too. Each successively lower taxonomical rank always has at least as many taxa as the higher levels, so you can’t simply count the total number of taxa at a rank. A single genus like Bifidobacterium, for example, can have dozens of species associated with it. For this reason, microbiologists usually measure diversity at the Family level: it’s a good compromise between overall coverage and specificity of taxa.
Other ways to measure diversity
Microbiome scientists use other diversity metrics as well. The Shannon Index borrows from Information Theory to ask how much unique information is contained in a given sample. A radio channel of random static, for example, would have a lower Shannon number than one for a music concert; random noise carries less information than a symphony. Similarly, a microbiome with a boring makeup — all the same species, for example — would have a lower Shannon number than one containing a rich abundance of many different types of microbes and abundances.
In practice, Shannon and Inverse Simpson tend to track one another reasonably well, a clue that they are getting at a similar idea. Shannon tends to fall within a narrower, more predictable range, so I prefer it over Inverse Simpson when looking at my own samples, but it often doesn’t matter which metric you use as long as you’re consistent.
In the real world, the type and variety of microbes in the body are constantly changing, so it’s important not to get too hung up on the number you got from a single sample. For example, here’s how my diversity changes day-to-day over the course of a month:
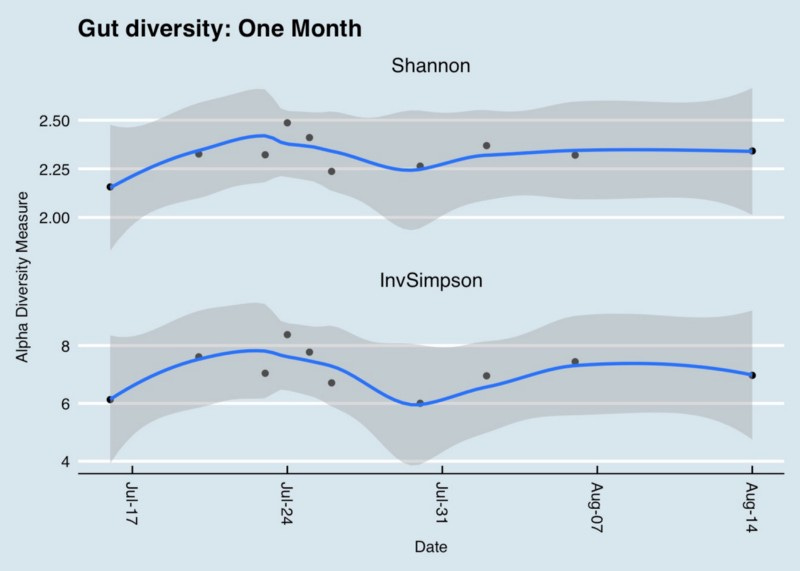
How two different diversity metrics compare over one month of my daily gut samples. Day-to-day variability is high, making it unreliable to consider a single result when deciding whether a given sample is “diverse”.
As you can see, there can be significant swings day-to-day. When people ask me how to fix their low microbiome diversity, my first suggestion is to test again. They may simply have picked the wrong day for the test!
The bottom line is that I have learned to not place much stock in any diversity measure. After all, whether diversity is “good” or “bad” depends on what is in the sample. Is high diversity good even if it includes many known pathogens? Is “low” diversity good if it only includes one or two known commensal bacteria?
As always in the microbiome world, it’s hard to tell.
Originally published at richardsprague.com on May 10, 2018.
